Blog (en-GB)
- Detalji
- Kategorija: Blog (en-GB)
Dr Olga Vasieva (transliteration used in earlier papers: Vassieva, born Potapova) https://www.researchgate.net/profile/Olga_Vasieva2
Dr Olga Vasieva is a director of the analytical company Ingenet. Ltd and an honorary lecturer at the University of Liverpool. Biophysicist by education, she had an experience in a number of experimental and computational areas of biology during her appointments at 5 distinguished academic centers in Russia, USA and UK (Institute of Theoretical and Experimental Biophysics, Moscow University, Georgetown University, Dundee University, Liverpool University) and 3 biotechnology/biomedicine companies (Integrated genomics Inc, USA, CXR Biosciences, UK, and Ingenet Ltd, UK). She has authored more than 60 scientific publications in peer-reviewed journals including high-impact journals such as PLOS ONE, Scientific reports, BMC genomics, Nuclear Acid research, and 2 book chapters. The work that she authored and co-authored has been cited more than 10,000 times (h-index 21, i10-29 (Google Scholar)). As a scientific supervisor she has trained 5 PhD and 9 MS students.
Dr Vasieva is an acknowledged reviewer for a number of high impact international scientific journals including Nuclear acid research, Bioinformatics (Oxford University Press), Systems biology in Reproductive medicine, Autophagy, Methods, Data mining and a number of Elsevier Press specialised journals. During the last 5 years she was invited as an expert to evaluate project proposals for the main UK funding bodies, BBSRC and MRC (Systems medicine) and also for EU international funding scheme ERACoSysMed (systems medicine) in 2017 and 2019 as an invited chair. She was a member of the data management group for the ERASYSBIO funding scheme, where she also took part as one of the successful Co-PIs (BBSRC-N 77093, GRAPPLE project). Dr Vasieva was also a Co-I in two NERC-funded collaborative projects (N 108470, N 141016) and, earlier in her career (2007), had received the TMRC systems medicine award.
Dr Vasieva is one of the pionere members of the international Fellowship of Interpretation of Genomes (FIG) and took part in the development of the SEED/RAST database. Moving in the systems medicine field, she has introduced and supported systems biology/medicine approaches in the Biomedical Research Centre at the University of Dundee and in the Institute of Integrative Biology (University of Liverpool) and established systems biology/bioinformatics consultancy and analytics company Ingenet Ltd (www.Ingenets.com). The latter is known for its novel approaches to data mining and analysis, and is collaborating with a number of scientific groups in UK and internationally (Okinawa University). The main research interests of Dr Vasieva include complex biological systems analysis, evolutionary and functional properties of metabolic pathways involved in cognition, cancer and reproduction, systems medicine and symbiotic metabolism in bacterial communities.
Dr Vasieva’s publications over the last 5 years:
|
1. Dr Vasieva’s publications over the last 5 years: Maclean, A, Bhullar, J, Miwa, S, von Glinicki, T, Morais, CLM, Martin, F, Valentijn, A, Vasieva, O, Tempest, N. Drury, J, Hapangama, D. Differential expression of hormone metabolising enzymes defines endometrial epithelial sub-populations. BLOG, 2019,126 (6): E134 . ISSN: 1470-0328 2. Maclean, A, Bhullar, J, Miwa, S, von Glinicki, T, Medeiros-de-Morais, CD, Martin, FL, Valentijn, AJ, Vasieva, O, Tempest, N, Drury, J, Hapangama, D. Multi-Methodological Approach Defines Sub-Populations of Epithelial Cells in Human Endometrium, 2019, Reproductive sciences, 26:103A, ISSN: 1933-7191 3. Vasieva O, Goryanin I, Is there a Function for a Sex Pheromone Precursor? Journal of integrative bioinformatics, 2019, doi:10.1515/jib-2019-0016 4. O Vasieva, S Cetiner, A Savage, GG Schumann, VJ Bubb, JP Quinn. Potential impact of primate-specific SVA retrotransposons during the evolution of human cognitive function Trends in Evolutionary Biology, 2017, 6 (1), doi:10.4081/ni.2017.6514 5. O Vasieva, A Sorokin, M Murzabaev, P Babiak, I Goryanin. A Study on the Analysis of Personal Gut Microbiomes, 2019, J Comput Sci Syst Biol 12, 71-79 6. KT Yee, O Vasieva, P Rojnuckarin, RNA-Sequencing of Snake Venom Glands, in Snake and Spider Toxins, January 2020, Springer, 87-96, doi:10.1007/978-1-4939-9845-6_5 7. Huang, F, Vasieva, O, Sun, YQ, Faulkner, M, Dykes, GF, Zhao, ZY, Liu, LN, Roles of RbcX in Carboxysome Biosynthesis in the Cyanobacterium Synechococcus elongatus PCC7942, 2019, Plant Physiol, 179 (1):184-194, doi: 10.1104/pp.18.01217 8. King, K. C., Brockhurst, M. A., Vasieva, O. et al, (2016). Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME JOURNAL, 10(8), 1915. doi: 10.1038/ismej 9. Shanmugasundram, A., Gonzalez-Galarza, F.F., Wastling, J. M., Vasieva, O., & Jones, A.R. (2014). An integrated approach to understand apicomplexan metabolism from their genomes. BMC BIOINFORMATICS, 15: S3-A3, doi:10.1186/1471-2105-15-S3-A3 10. Mueller, A, Tew, S, R, Vasieva, O, Clegg PD, Canty-Laird E, A systems biology approach to defining regulatory mechanisms for cartilage and tendon cell phenotypes. Scientific reports. 2016, 1-14.doi: 10.1038/srep33956 11. Bell, R, Barroclough, R, Vasieva O. Gene expression meta-analysis of potential metastatic breast cancer markers. Current Molecular Medicine. 2017, 17: 1-10 doi:10.2174/1566524017666170807144946 12. Stenslokken, K. -O., Ellefsen, S., Vasieva, O. et al, Life without Oxygen: Gene Regulatory Responses of the Crucian Carp (Carassius carassius) Heart Subjected to Chronic Anoxia. PlosOne. 2014, 9(11), doi:10.1371/journal.pone.0109978 13. Mathew, D., Drury, J. A., Valentijn, A. J.,Vasieva, O., & Hapangama, D. K. Insilico, in vitro and in vivo analysis identifies a potential role for steroid hormone regulation of FOXD3 in endometriosis-associated genes. HUMAN REPRODUCTION, 2016, 31(2), 345-354, doi:10.1093/humrep/dev307 14. N Mirza, O Vasieva, R Appleton, S Burn, D Carr, D Crooks, D du Plessis, Duncan, R, Farah, JO. Josan, V, Miyajima, F, Mohanraj, R, Shukralla, A, Sills, GJ, Marson, AG, Pirmohamed, M. An integrative in silico system for predicting dysregulated genes in the human epileptic focus: Application to SLC transporters, 2016, Epilepsia 57 (9), 1467-1474, doi:10.1111/epi.13473 15. Capece, A, Vasieva, O, Meher, S, Alfirevic Z, Alfirevic A. Pathway Analysis of Genetic Factors Associated with Spontaneous Preterm Birth and Pre-Labor Preterm Rupture of Membranes. PlosOne. 2014, 9(9):e108578, doi: 10.1371/journal.pone.0108578 16. Kulohoma BW, Marriage F, Vasieva O, Enitan D, CarrolPeripheral blood RNA gene expression in children with pneumococcal meningitis: a prospective case– control study, 2017, BMJ Paediatrics Open, 2017, 1:e000092. doi:10.1136/bmjpo-2017-000092 17. Marriott, A. S., Vasieva, O., Fang, Y. et al. NUDT2 Disruption Elevates Diadenosine Tetraphosphate (Ap(4)A) and Down-Regulates Immune Response and Cancer Promotion Genes. PlosOne. 2016. 11(5), doi:10.1371/journal.pone.0154674 18. R Eakins, J Walsh, L Randle, RE Jenkins, I Schuppe-Koistinen, C Rowe, .Olga Vasieva, Prats, N, Brillant, N, Auli, M, Bayliss, M, Webb, S, Rees, JA, Kitteringham, NR, Goldring, CE, Park, BK, Adaptation to acetaminophen exposure elicits major changes in expression and distribution of the hepatic proteome..Scientific reports, 2015, 5, 16423, doi:10.1038/srep16423 19. M Sedic, LA Gethings, JPC Vissers, JP Shockcor, S McDonald, O Vasieva, Lemac, M, Langridge, JI, Batinic, D, Pavelic, SK. Label-free mass spectrometric profiling of urinary proteins and metabolites from paediatric idiopathic nephrotic syndrome, ...Biochemical and biophysical research communications 2014, 452 (1), 21-26, doi:10.1016/j.bbrc.2014.08.016 20. A Warburton, O Vasieva, P Quinn, JP Stewart, JP Quinn, Statistical analysis of human microarray data shows that dietary intervention with n-3 fatty acids, flavonoids and resveratrol enriches for immune response and …,British Journal of Nutrition, 2018, 119 (3), 239-249, doi:10.1017/S0007114517003506 21. D Avraam, S Arnold, O Vasieva, B Vasiev, On the heterogeneity of human populations as reflected by mortality dynamics, 2018, Aging (Albany NY) 8 (11), 3045, doi: 10.18632/aging.101112 22. Khilyas, I, Sorokin, A, Vassieva, O, Cohen, M, Goryanin, I, Two-component regulation systems of dominated electrogenic bacteria in microbial fuel cells treating swine wastewater, FEBS OPEN BIO Meeting Abstract: ShT.06-2 2018, 8: 43-43 Supplement: 1 Published: JUL 2018 23. V Yip, E Zhang, K Clarke, B Francis, L Rainbow, F Falciani, O Vasieva, Park, K, Naisbitt, D. Pirmohamed, M, Assessment of gene expression profiles of peripheral blood mononuclear cells from patients with a history of carbamazepine hypersensitivity, The Lancet, 389, S104. ISSN: 0140-6736. 24. Yee, KT, Tongsima, S, Vasieva, O, Ngamphiw, C, Wilantho, A, Wilkinson, MC, Somparn, P, Pisitkun, T, Rojnuckarin, P, Analysis of snake venom metalloproteinases from Myanmar Russell's viper transcriptome, TOXICON, 2018, 146: 31-41 doi:10.1016/j.toxicon.2018.03.005 25. Mathew, D, Drury, JA, Valentijn, AJ, Vasieva, O, Hapangama, DK, In silico, in vitro and in vivo analysis identifies a potential role for steroid hormone regulation of FOXD3 in endometriosis-associated genes, HUMAN REPRODUCTION, 2016, 31(2): 345-354 doi: 10.1093/humrep/dev307 26. Valba, OV, Nechaev, SK, Sterken, MG, Snoek, LB, Kammenga, JE, Vasieva, OO, On predicting regulatory genes by analysis of functional networks in C. elegans, BIODATA MINING, 2015, 8:33 doi:10.1186/s13040-015-0066-0 27. Huxley M, Stead J, Vasieva O. Streptococcal infection-related autoimmunity and autism: crosstalk in protein functional networks, J. of Systems and Integrated Neuroscience, 2017, 3(4): 1-10, doi:10.15761/JSIN.1000171 28. Shanmugasundram A, Gonzalez-Galarza FF, Wastling JM, Vasieva O, Jones A, An integrated approach to understand apicomplexan metabolism from their genomes, BMC Bioinformatics, 2014, 15 (Suppl 3):A3, doi:10.1186/1471-2105-15-S3-A3 |
- Detalji
- Kategorija: Blog (en-GB)
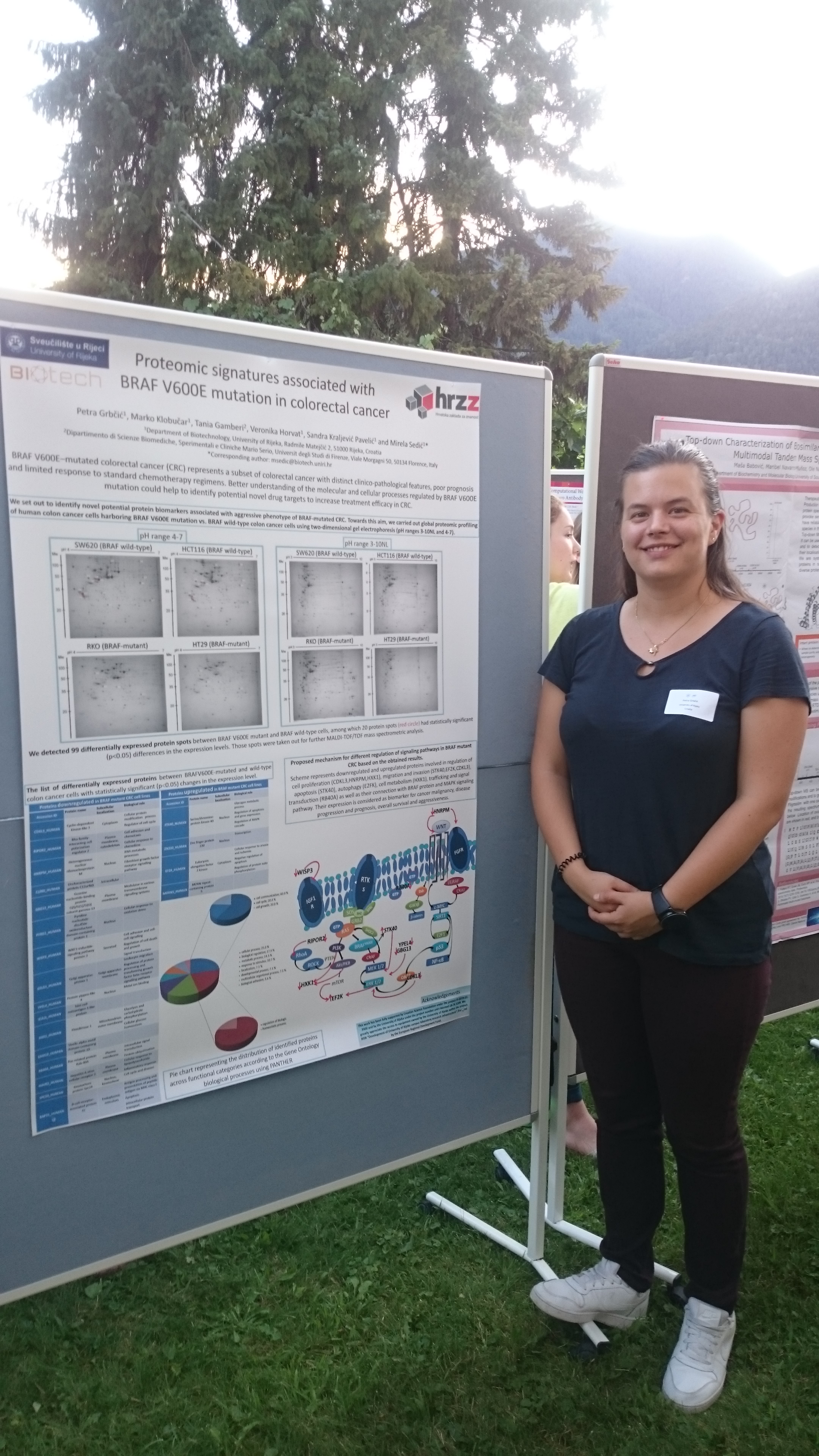
FEBS 2019 Advanced course on Advanced proteomics was held in Varna, Italy from 28. July - 3. August 2019.

This summer school provided graduate students and young postdoctoral scientists from academia and industry with insights into state-of-the-art proteomic technologies and applications in the life sciences.
PhD student Petra Grbčić held an poster presentation : "Proteomic signatures associated with BRAF V600E mutation in colorectal cancer"
- Detalji
- Kategorija: Blog (en-GB)
Associate Professor Mirela Sedić, PhD, University of Rijeka, Department of Biotechnology, Croatia
Dr Olga Vasieva, University of Liverpool/Ingenet. Ltd., UK
Dr Thomas Eichmann, University of Graz Institute of Molecular Biosciences, Graz, Austria
Dr Marko Klobučar, University of Rijeka, Department of Biotechnology, Croatia
Associate Professor Elitza Markova Car, PhD, University of Rijeka, Department of Biotechnology, Croatia
Full Professor Djuro Josić, PhD, University of Rijeka, Department of Biotechnology, Croatia
Petra Grbčić, mag. pharm. inv., University of Rijeka, Department of Biotechnology, Croatia
Iris Car, mag. biotech. in med., University of Rijeka, Department of Biotechnology, Croatia
Associate Professor, Sandra Kraljević Pavelić, PhD, University of Rijeka, Department of Biotechnology, Croatia
- Detalji
- Kategorija: Blog (en-GB)
2021
1. 11th International Ceramide Conference 19 - 22 April 2021 (VIRTUAL),
Petra Grbčić, Thomas Eichmann, Sandra Kraljević Pavelić, Mirela Sedić
Altered sphingosine-1-phosphate/ceramide balance contributes to the development of resistance to vemurafenib in BRAFV600E mutant colon cancer cells
2. EACR 2021 Congress, Innovative Cancer Science: Better Outcomes through Research held virtually from 09 - 12 June 2021
M. Sedić*, I. Car*, A. Dittmann, O. Vasieva, P. Grbčić, M. Klobučar, S. Kraljević Pavelić. (* equal contribution)
Integrated proteomics and bioinformatics approach reveals caveolae-associated signalling as novel druggable vulnerability of vemurafenib-resistant colon cancer cells with BRAFV600E mutation - poster presentation
3. 2nd International Conference on Cell and Experimental Biology (CEB-2021) JULY 12-14, 2021 | VIRTUAL; invited talk
M. Sedić
"Proteomic Profiling of BRAFV600E Mutant Colon Cancer Cells Reveals the Role of Nucleophosmin in Mediating the Resistance to BRAF Inhibition by Vemurafenib"
2019
1. CEOC 2019, 15th Central European Oncology Congress, Opatija, Croatia
Petra Grbčić, Sandra Kraljević Pavelić, Mirela Sedić
Differences in molecular and cellular characteristics between BRAF V600E-mutated vemurafenib resistant and sensitive colon cancer cell lines
Iris Car, Marko Klobučar, Sandra Kraljević Pavelić, Mirela Sedić
Proteomic profiling of BRAF V600E mutant colon cancer cells with acquired resistance to vemurafenib
2. 13th Mass Spectrometry School in Biotechnology and Medicine (MSBM) - Dubrovnik, Croatia
Iris Car, Marko Klobučar, Sandra Kraljević Pavelić, Mirela Sedić
Optimization of protocol for secretome collection from vemurafenib-resistant BRAFV600E mutant colon cancer cells for mass spectrometry-based proteome analysis
3. 13th European Summer School on Advanced Proteomics, FEBS 2019 Advanced Course, Varna, Italy
Petra Grbčić, Marko Klobučar, Tania Gamberi, Sandra Kraljević Pavelić, Veronika Horvat, Mirela Sedić
Proteomic signatures associated with BRAF V600E mutation in colorectal cancer
4. International Doctoral Conference, AARC, University of Rijeka, Croatia
Iris Car, Marko Klobučar, Sandra Kraljević Pavelić, Mirela Sedić
Secretome analysis of BRAF V600E-mutant colon cancer cells with acquired resistance to vemurafenib
Petra Grbčić, Sandra Kraljević Pavelić, Mirela Sedić
Sphingolipid metabolism is dysregulated in BRAF V600E mutant colon cancer cells resistant to vemurafenib
Stranica 4 od 5

